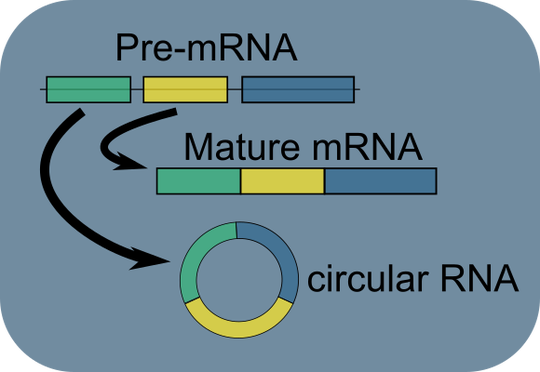
Our lab focuses on the expression, splicing, and regulatory roles of coding and non-coding RNAs, such as circular RNAs, in cardiovascular and stress-related systems.
Our research integrates high-throughput sequencing, transcriptomics, and systems biology to uncover how cells respond and adapt at the molecular level.
We are actively involved in open-source software development, creating tools that support reproducible and scalable analyses in RNA research.

The Jakobi Lab is part of the Translational Cardiovascular Research Center (TCRC) in the Department of Internal Medicine at the The University of Arizona College of Medicine – Phoenix.

Join Our Team!
We are looking for a Postdoctoral Research Associate, Master’s and PhD students, and a Scientific Analyst.
News
We are delighted to introduce Walker Mellon, our newest team member at the TCRC! Walker is a data scientist with expertise in computational models, data analytics, bioinformatics, and phylogenetics. He has a strong background in programming languages such as Python and R and will be playing a key role in advancing our NIH-funded projects.
Welcome to the team, Walker!
Tobias’ invited editorial “Translating Translation: Protein-Coding CircRNAs as Potential Therapeutical Targets in Coronary Artery Disease” in JACC: Basic to Translational Science has been published! The editorial comments on the recent study “CircBTBD7-420aa Encoded by hsa_circ_0000563 Regulates the Progression of Atherosclerosis and Construction of circBTBD7-420aa Engineered Exosomes” by Gan et al. and highlights the translation of circBTBD7 into a functional peptide that can regulate the progression of atherosclerosis. The editorial also discusses techniques and approaches required to validate translation of circRNAs based on computational predictions.
Are you ready to revolutionize our understanding of circular RNAs (circRNAs) and their role in heart disease? We’re seeking a highly motivated and talented computational biologist to join our team at the Jakobilab!
Be Part of a Cutting-Edge Research Environment
As a postdoctoral researcher in our lab, you’ll have the opportunity to contribute to the development of new features for circtools, our widely-used circRNA analysis software. You’ll work alongside world-class researchers and scientists who are passionate about advancing the field of circRNA research.
Responsibilities:
- Design, implement, and validate novel algorithms and tools for analyzing circRNA expression, function, and interactions
- Collaborate with experimentalists to integrate computational findings with wet-lab experiments
- Analyze large-scale RNA sequencing datasets to identify key circRNAs involved in heart disease pathogenesis
- Contribute to the development of new features for circtools
- Disseminate research through publications and presentations at scientific conferences
Requirements:
- PhD in Bioinformatics, Computational Biology, Computer Science, or a related field
- Expertise in programming languages such as Python, R, or other relevant languages
- Experience with next-generation sequencing data analysis and RNA biology
- Strong communication and teamwork skills
- A passion for pushing the boundaries of scientific discovery
Why Join Our Lab?
Our lab is committed to fostering a collaborative and inclusive research environment that values innovation, creativity, and teamwork. As a postdoctoral researcher in our lab, you’ll have access to cutting-edge resources, state-of-the-art facilities, and a supportive team of researchers who are dedicated to advancing the field of circRNA research.
How to Apply:
If you’re ready to join our dynamic team, please submit your CV and a brief statement of research interests via our application portal. We look forward to hearing from you!
We are proud to release circtools 2.0 on GitHub and the accompanying paper on bioRxiv in collaboration with the Dieterich Lab.
Circtools 2.0 features 4 completely new modules for computational circular RNA research:
- conservation module: enables users to perform circRNA conservation analysis in five widely studied animal model species: mouse, human, rat, pig, and dog.
- padlock probe design: a specialized primer design tool tailored specifically for next-generation spatial transcriptomics technology.
- nanopore module: requires sequencing reads that have been produced using the protocol outlined in Rahimi et al. (2021) and enables full-length circRNA detection.
- metatool module: offers integration of circularRNA predictions from CIRIquant to boost recall rates for circRNA detection.
Research
Teaching
Principal Investigator
Dr. Tobias Jakobi is a computational biologist whose research focuses on understanding the expression, splicing, and interaction of coding and non-coding RNAs, such as circular RNAs, particularly in the context of cardiovascular and stress-related physiology. His work bridges high-throughput sequencing, transcriptomics, and systems biology, offering powerful insights into how cells adapt at the molecular level.
I am working in the Department of Internal Medicine and in the new Translational Cardiovascular Research Center (TCRC) at The University of Arizona College of Medicine – Phoenix .
- Computational RNA biology
- Bioinformatics
- Networks and functions of RNAs in the heart
- Systems cardiology
- High performance computing
-
Dr. rer. nat. in Bioinformatics, 2014
Bielefeld University, Germany
-
MSc in Bioinformatics & Genome Research, 2009
Bielefeld University, Germany
-
BSc in Bioinformatics & Genome Research, 2007
Bielefeld University, Germany
Selected publications
Bluesky
Contact
- tjakobi [at] arizona [dot] edu
- 475 N 5th St, Phoenix, AZ 85004
- Please register in the Biomedical Sciences Partnership Building (BSPB)
- @Jakobilab on Bluesky